Supplemental Data: Difference between revisions
Mmaciejewski (talk | contribs) Created page with "= Supplemental Data = NAN enables users to enrich archived datasets with additional context and information through the use of supplemental data. While NDTS harvests all files present in an experimental dataset—including waveform files, pre-compiled pulse programs, and other required components—certain files and metadata relevant to downstream use may not be available at the time of data collection. These may include: * Protocols * Data processing and analysis scri..." |
Mmaciejewski (talk | contribs) No edit summary |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Datasets}} | |||
== Overview == | |||
NAN enables users to enrich archived datasets with additional context and information through the use of supplemental data. While NDTS harvests all files present in an experimental dataset—including waveform files, pre-compiled pulse programs, and other required components—certain files and metadata relevant to downstream use may not be available at the time of data collection. | NAN enables users to enrich archived datasets with additional context and information through the use of supplemental data. While NDTS harvests all files present in an experimental dataset—including waveform files, pre-compiled pulse programs, and other required components—certain files and metadata relevant to downstream use may not be available at the time of data collection. | ||
These may include: | These may include: | ||
* Protocols | * Protocols | ||
* | * Scripts | ||
* | * Identifiers | ||
* | * Files | ||
== Adding Supplemental Data == | == Adding Supplemental Data == | ||
[[File:Supplemental Data Modal.png|thumb|Supplemental Data Modal|600x600px]] | |||
Supplemental data may be appended to individual datasets or dataset collections via a dedicated modal launched from the Dataset Editor or the dataset context menu | Supplemental data may be appended to individual datasets or dataset collections via a dedicated modal launched from the Dataset Editor or the dataset context menu. Entries in the supplemental data table require the following fields: | ||
* '''Category''' | * '''Category''' | ||
| Line 17: | Line 18: | ||
* '''Value''' | * '''Value''' | ||
* '''(Optional) Description''' | * '''(Optional) Description''' | ||
'''Types''' include: | |||
* URLs | * URLs | ||
* File uploads | * File uploads | ||
| Line 24: | Line 24: | ||
* External ID numbers | * External ID numbers | ||
'''Categories''' include: | |||
* '''Protocols''' – Instrument setup, sample preparation, pulse program setup, signal processing, and analysis | * '''Protocols''' – Instrument setup, sample preparation, pulse program setup, signal processing, and analysis | ||
* '''Scripts''' – Experimental setup, signal processing, and analysis | * '''Scripts''' – Experimental setup, signal processing, and analysis | ||
* '''Identifiers''' – BMRB, PDB, Metabolomics Workbench, DOIs | * '''Identifiers''' – BMRB, PDB, Metabolomics Workbench, DOIs | ||
* '''Data Files''' – Sample schedules, experimental parameters, pulse programs, processed data, sample parameters | * '''Data Files''' – Sample schedules, experimental parameters, pulse programs, processed data, sample parameters | ||
'''Values''' are user entered and must conform to the type. For example, URLs must be valid, BMRB and PDB IDs must be in the correct format, etc. | |||
The '''description''' field is optional | |||
== Supplemental Data in the Data Browser == | == Supplemental Data in the Data Browser == | ||
| Line 37: | Line 38: | ||
* A ''Supplemental Data'' column in the browser table supports filtering and sorting based on presence of supplemental data. | * A ''Supplemental Data'' column in the browser table supports filtering and sorting based on presence of supplemental data. | ||
== | == Supplemental Data via Download == | ||
Supplemental data entries become part of the dataset record, and any uploaded files are stored in a dedicated `supplemental_data` directory within the dataset. These entries and files are included in the downloadable dataset archive. In addition, a supplemental_data.csv file is stored in the dataset with a full list of all supplemental data entries. See the [[Download Datasets|Downloads]] section for full details. | |||
Latest revision as of 17:06, 10 June 2025
Overview
NAN enables users to enrich archived datasets with additional context and information through the use of supplemental data. While NDTS harvests all files present in an experimental dataset—including waveform files, pre-compiled pulse programs, and other required components—certain files and metadata relevant to downstream use may not be available at the time of data collection.
These may include:
- Protocols
- Scripts
- Identifiers
- Files
Adding Supplemental Data
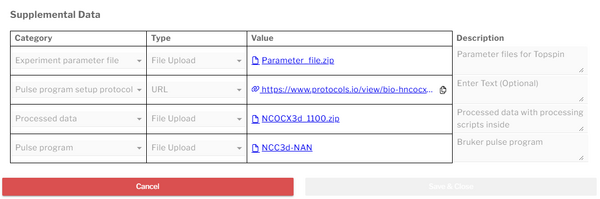
Supplemental data may be appended to individual datasets or dataset collections via a dedicated modal launched from the Dataset Editor or the dataset context menu. Entries in the supplemental data table require the following fields:
- Category
- Type
- Value
- (Optional) Description
Types include:
- URLs
- File uploads
- Existing files within the dataset
- External ID numbers
Categories include:
- Protocols – Instrument setup, sample preparation, pulse program setup, signal processing, and analysis
- Scripts – Experimental setup, signal processing, and analysis
- Identifiers – BMRB, PDB, Metabolomics Workbench, DOIs
- Data Files – Sample schedules, experimental parameters, pulse programs, processed data, sample parameters
Values are user entered and must conform to the type. For example, URLs must be valid, BMRB and PDB IDs must be in the correct format, etc.
The description field is optional
Supplemental Data in the Data Browser
- Datasets with supplemental data are marked with a special icon badge.
- A Supplemental Data column in the browser table supports filtering and sorting based on presence of supplemental data.
Supplemental Data via Download
Supplemental data entries become part of the dataset record, and any uploaded files are stored in a dedicated `supplemental_data` directory within the dataset. These entries and files are included in the downloadable dataset archive. In addition, a supplemental_data.csv file is stored in the dataset with a full list of all supplemental data entries. See the Downloads section for full details.