New Sample Creation: Difference between revisions
Apozhidaeva (talk | contribs) No edit summary |
Apozhidaeva (talk | contribs) No edit summary |
||
| (48 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<span style="display:inline-block; margin-bottom:1em;">[[Samples|← Back to Sample Browser ]]</span> | |||
{{DISPLAYTITLE: Creating/Cloning Samples}} | {{DISPLAYTITLE: Creating/Cloning Samples}} | ||
[[File:New Sample.png|thumb|487x487px|Creating a new sample]] | |||
=== Single sample creation === | === Single sample creation === | ||
To create a new sample | To create a new sample | ||
| Line 10: | Line 13: | ||
# After completing the required fields, four additional tabs become editable, allowing you to specify further sample details including: | # After completing the required fields, four additional tabs become editable, allowing you to specify further sample details including: | ||
#* '''Container:''' specify details about tube, rotor or other container holding the sample | #* '''Container:''' specify details about tube, rotor or other container holding the sample | ||
#* '''Composition:''' create a list of components with details such as concentration and isotope labeling for each of the constituents | #* '''Composition:''' create a list of [[Sample Components|components]] with details such as concentration and isotope labeling for each of the constituents. Additional component-specific information can be added by pressing the '''“'''Add Component Details'''”''' (pencil icon) under ''Actions''. | ||
#* '''Sample Properties:''' enter biophysical properties of the sample | #* '''Sample Properties:''' enter biophysical properties of the sample | ||
#* '''Additional Info:''' provide notes for a facility about how to store and handle your sample before, between and after experiments | #* '''Additional Info:''' provide notes for a facility about how to store and handle your sample before, between and after experiments | ||
#* Please note that each tab also has "'''Notes'''" section where any additional info can be entered | #* Please note that each tab also has "'''Notes'''" section where any additional info can be entered | ||
# Enter information about molecular interactions (Optional). If you have two or more molecules added to the sample composition you can specify interaction details using '''Interaction Manager.''' Note, that you need to save the sample after completing its composition for ''' | #* ''Detailed description of all options can be found in the [[#New_Sample|table]] below.'' | ||
# Press "Save and Close" button once you are done. Once the sample was saved it will appear in Samples section of the Data Browser | # Enter information about molecular interactions (Optional). If you have two or more molecules added to the sample composition you can specify interaction details using '''Interaction Manager.''' Note, that you need to save the sample after completing its composition for Interaction Manager '''t'''o become active. | ||
# Press "Save and Close" button once you are done. Once the sample was saved it will appear in Samples section of the Data Browser. | |||
---- | |||
=== Interaction Manager === | |||
[[File:Screenshot 2025-07-17 at 3.55.53 PM.png|thumb|383x383px|Interaction Matrix of the Interaction Manager]] | |||
The Interaction Manager allows users to define known interactions between molecules in a sample. To access it: | |||
* Navigate to the Composition section. | |||
* Add '''at least two components''' to the sample | |||
* Press '''Save''' to enable the Interaction Manager button (it will appear green and active in the top-right corner of the components table). | |||
Clicking on the Interaction manager opens an interaction matrix displaying all possible '''pairwise interactions''' between the components. Clicking on a square for the interaction of interest opens a dialog box where the following information can be entered: | |||
*'''Interaction Type''': choose between covalent or non-covalent | |||
*'''Stoichiometry''': enter the ratio of the interacting components | |||
*'''Binding Affinity''': specify whether the affinity is ''greater than'', ''less than'', or ''equal to'' a desired value. | |||
---- | |||
=== Creating multiple samples === | === Creating multiple samples === | ||
| Line 30: | Line 49: | ||
# After completing the required fields, four additional tabs become editable, allowing you to specify further details for the <u>first sample</u> including: | # After completing the required fields, four additional tabs become editable, allowing you to specify further details for the <u>first sample</u> including: | ||
#* '''Container:''' specify details about tube, rotor or other container holding the sample | #* '''Container:''' specify details about tube, rotor or other container holding the sample | ||
#* '''Composition:''' create a list of components with details such as concentration and isotope labeling for each of the constituents | #* '''Composition:''' create a list of [[Sample Components|components]] with details such as concentration and isotope labeling for each of the constituents | ||
#* '''Sample Properties:''' enter biophysical properties of the sample | #* '''Sample Properties:''' enter biophysical properties of the sample | ||
#* '''Additional Info:''' store any other details you wish to associate with the sample | #* '''Additional Info:''' store any other details you wish to associate with the sample | ||
#* Please note that each tab also has "'''Notes'''" section where any additional info can be entered | #* Please note that each tab also has "'''Notes'''" section where any additional info can be entered | ||
'''IMPORTANT NOTE''': ''before proceeding to the next step, make sure you fill all the required information about the first sample.'' | |||
# Select one or more fields that will vary between the samples. The selected fields will become highlighted | <li value="6"> Once the information about the first sample is complete, click the '''"Switch to Variable Field Selection"''' button located in the top-right corner of the sample creation window | ||
# Select one or more fields that will vary between the samples. The selected fields will become highlighted in '''''blue''''' | |||
# Click "Close and Continue" button to open '''Milti-Sample Creation Wizard''' where you are required to provide a name for the Sample Collection and a number of samples to be created. | # Click "Close and Continue" button to open '''Milti-Sample Creation Wizard''' where you are required to provide a name for the Sample Collection and a number of samples to be created. | ||
# Click the "Create N Sample Rows" (N is the specified number of samples) button and enter the varying information for each sample in the displayed table. | # Click the "Create N Sample Rows" (N is the specified number of samples) button and enter the varying information for each sample in the displayed table. | ||
| Line 43: | Line 63: | ||
#* import the CSV file to populate the rows for the variable fields | #* import the CSV file to populate the rows for the variable fields | ||
# Press "Create Samples" button to save the sample collection. Once saved, the sample collection will Appear in Spawned Sample Collections section of the Data Browser | # Press "Create Samples" button to save the sample collection. Once saved, the sample collection will Appear in Spawned Sample Collections section of the Data Browser | ||
---- | |||
=== Cloning a sample === | === Cloning a sample === | ||
'''Cloning a sample''' | '''Cloning a sample''' feature allows you to quickly create a new sample by copying information from the existing sample and making only the necessary modifications. To do so | ||
# Go to | # Go to Data Browser → Samples | ||
# | # Locate the sample you would like to clone | ||
# Enter a name for the new sample | # Right-click on the sample and choose "Clone" option | ||
# | # Enter a unique name for the new sample in the popup window | ||
# The '''Edit Sample''' window will appear. Make any necessary changes to the new sample by editing information in any of the five tabs: | |||
#* Sample | #* Sample | ||
#* Container | #* Container | ||
| Line 54: | Line 77: | ||
#* Sample Properties | #* Sample Properties | ||
#* Additional Info | #* Additional Info | ||
# Click Save and Close button to save the sample. The new sample will appear in the Samples section of the Data Browser. | # Click "Save and Close" button to save the sample. The new sample will appear in the Samples section of the Data Browser. | ||
{| class="wikitable" | |||
|+<span id="New_Sample">New Sample</span> | |||
!Tab | |||
!Name | |||
!Description | |||
|- | |||
| rowspan="8" |Sample | |||
|'''PI (required)''' | |||
|User selected PI (consists of PI(s) linked to the user | |||
|- | |||
|'''Display Name (required)''' | |||
|User specified name | |||
|- | |||
|'''Physical State (required)''' | |||
|Solid-state NMR or solution NMR | |||
|- | |||
|Preparation Date | |||
|Date when sample was prepared | |||
|- | |||
|Preparation URL | |||
|Link to paper/protocols.io, etc. | |||
|- | |||
|Description | |||
|Detailed description of the sample | |||
|- | |||
|Notes | |||
|Other relevant sample information that the user wishes to enter | |||
|- | |||
|Tags | |||
|List of user-defined tags that can be used for search and filter | |||
|- | |||
| rowspan="2" |Container | |||
|Container Type | |||
|Information about rotor, tube or. other container | |||
|- | |||
|Notes | |||
|Other relevant information related to the sample container that the user wishes to enter | |||
|- | |||
|Composition | |||
|Type | |||
|[[Sample Components|Sample Components]] | |||
|- | |||
| | |||
|Name | |||
|User defined name of the pocmponent | |||
|- | |||
| | |||
|Role | |||
|Role of the component. The following options are available: | |||
* Molecule of interest | |||
* Solvent | |||
* Buffer | |||
* Salt | |||
* Chemical Shift Standard | |||
* Detergent | |||
* Excipient | |||
* Polarizing Agent | |||
* Paramagnetic Agent | |||
* Alignment Media | |||
* Membrane mimetic | |||
* Interacts with molecule of interest | |||
* Other | |||
|- | |||
| | |||
|Quantity | |||
|User defined quantity of the component | |||
|- | |||
| | |||
|Unit | |||
|'''Concentration''' | |||
* mM | |||
* μM | |||
* M | |||
* mg/mL | |||
* g/mL | |||
* v/v % | |||
* N | |||
'''Mass''' | |||
* µg | |||
* ng | |||
* mg | |||
* g | |||
'''Volume''' | |||
* mL | |||
* µL | |||
* cc<sup>3</sup> (note: display as CC<sup>3</sup> on front end, write as CC^3 to back end) | |||
|- | |||
| | |||
|Isotopic Labeling | |||
|Following options are available: | |||
* U-15N | |||
* U-13C | |||
* U-2H | |||
* U-15N-U-13C | |||
* U-15N-U-13C-U-2H | |||
* Natural Abundance | |||
* Selective | |||
* Site-specific | |||
Additional user defined isotopic labeling details can be added/modified in the '''Actions''' | |||
|- | |||
| | |||
|Actions | |||
|Following actions are available: | |||
*'''Remove''' the component | |||
*'''Edit component details''' (pencil icon) opens a dialog box where additional information can be added for each component.The available fields depend on the component type (for example, UniProt ID can be added for a protein and PUBChem ID for organic compounds} | |||
*'''Edit Isotopic Details''' (atom icon) allows to modify isotopic labeling. | |||
# If Isotope Labelling are U-15N, U-13C, U-2H, U-15N-U-13C, U-15N-U-13C-U-2H, there will be a predefined form with | |||
#* Isotope | |||
#* Enrichment % for Segment/Residue/Atom | |||
#* Metabolic Precursor | |||
#* Other | |||
# If Isotope Labelling are Natural Abundance, Selective, Site-specific, the user has to manually add isotope first. | |||
|- | |||
|Sample properties | |||
|pH Directly Measured | |||
|Indicate if the pH was measured directly in the NMR sample | |||
|- | |||
| | |||
|pH value | |||
|pH value | |||
|- | |||
| | |||
|Aligned | |||
|Specify if the sample is under anisotropic alignment conditions (used for RDC or CSA measurements). | |||
|- | |||
| | |||
|Paramagnetic | |||
|Indicate if the sample contains paramagnetic center(s) | |||
|- | |||
| | |||
|Conductivity measured | |||
|Indicate whether the electrical conductivity of the sample was experimentally measured. If so, the value can be provided in μS/cm | |||
|- | |||
| | |||
|Sample volume | |||
|Sample volume in μL | |||
|- | |||
| | |||
|Ionic strength | |||
|ionic strength of the sample in mM | |||
|- | |||
| | |||
|Notes | |||
|Other relevant sample information that the user wishes to enter | |||
|- | |||
|Additional Info | |||
|Storage/Handling details | |||
|User provided storage and handling conditions for the sample | |||
|- | |||
| | |||
|Notes | |||
|Other relevant sample information that the user wishes to enter | |||
|} | |||
[[Category:Data Browser]] | [[Category:Data Browser]] | ||
Latest revision as of 18:47, 21 August 2025
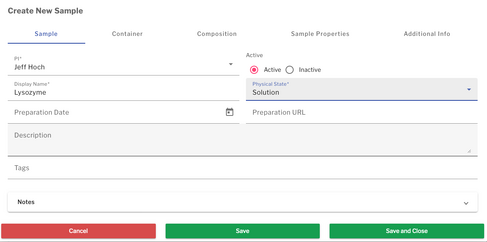
Single sample creation
To create a new sample
- Navigate to Samples page by going to Data Browser → Samples
- Click "Create a new sample" button located in the top-right part of the screen, above the table listing all the existing samples
- From the two options provided, choose Single Sample
- In the appeared window, fill general information in the Sample tab. Two fields are required (marked with an asterisk (*)) for each sample before it can be saved:
- Display Name: a unique identifier of the sample
- Physical State: whether the sample is in solution or solid form.
- After completing the required fields, four additional tabs become editable, allowing you to specify further sample details including:
- Container: specify details about tube, rotor or other container holding the sample
- Composition: create a list of components with details such as concentration and isotope labeling for each of the constituents. Additional component-specific information can be added by pressing the “Add Component Details” (pencil icon) under Actions.
- Sample Properties: enter biophysical properties of the sample
- Additional Info: provide notes for a facility about how to store and handle your sample before, between and after experiments
- Please note that each tab also has "Notes" section where any additional info can be entered
- Detailed description of all options can be found in the table below.
- Enter information about molecular interactions (Optional). If you have two or more molecules added to the sample composition you can specify interaction details using Interaction Manager. Note, that you need to save the sample after completing its composition for Interaction Manager to become active.
- Press "Save and Close" button once you are done. Once the sample was saved it will appear in Samples section of the Data Browser.
Interaction Manager
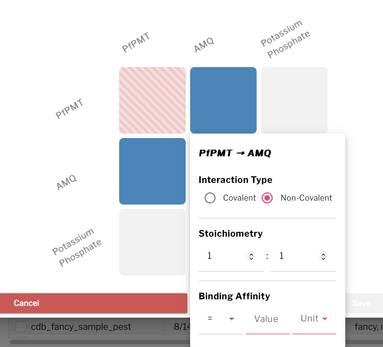
The Interaction Manager allows users to define known interactions between molecules in a sample. To access it:
- Navigate to the Composition section.
- Add at least two components to the sample
- Press Save to enable the Interaction Manager button (it will appear green and active in the top-right corner of the components table).
Clicking on the Interaction manager opens an interaction matrix displaying all possible pairwise interactions between the components. Clicking on a square for the interaction of interest opens a dialog box where the following information can be entered:
- Interaction Type: choose between covalent or non-covalent
- Stoichiometry: enter the ratio of the interacting components
- Binding Affinity: specify whether the affinity is greater than, less than, or equal to a desired value.
Creating multiple samples
Creating multiple samples feature is a convenient tool that allows batch creation of similar samples that vary by one or more parameters. Examples of such sample collections include but are not limited to:
- Titrations: multiple samples of molecule of interest at fixed concentration and varying concentration of a binding partner
- Screening samples: multiple samples of a molecule of interest with varying binding partners
Multiple samples created this way will be combined into a collection and can be edited together later. To create multiple samples:
- Navigate to Samples page by going to Data Browser → Samples
- Click "Create a new sample" button located in the top-right part of the screen, above the table listing all the existing samples
- From the two options provided, choose Multiple Samples
- In the appeared window, fill general information for the first sample in the Sample tab. Two fields are required (marked with an asterisk (*)) for each sample before it can be saved:
- Display Name: a unique identifier of the sample
- Physical State: whether the sample is in solution or solid form.
- After completing the required fields, four additional tabs become editable, allowing you to specify further details for the first sample including:
- Container: specify details about tube, rotor or other container holding the sample
- Composition: create a list of components with details such as concentration and isotope labeling for each of the constituents
- Sample Properties: enter biophysical properties of the sample
- Additional Info: store any other details you wish to associate with the sample
- Please note that each tab also has "Notes" section where any additional info can be entered
IMPORTANT NOTE: before proceeding to the next step, make sure you fill all the required information about the first sample.
- Select one or more fields that will vary between the samples. The selected fields will become highlighted in blue
- Click "Close and Continue" button to open Milti-Sample Creation Wizard where you are required to provide a name for the Sample Collection and a number of samples to be created.
- Click the "Create N Sample Rows" (N is the specified number of samples) button and enter the varying information for each sample in the displayed table.
- Alternatively, you can import data in CSV format. To do this
- download the template generated for multiple sample entry and edit it in any editor
- or format your own file to match the template structure
- import the CSV file to populate the rows for the variable fields
- Press "Create Samples" button to save the sample collection. Once saved, the sample collection will Appear in Spawned Sample Collections section of the Data Browser
Cloning a sample
Cloning a sample feature allows you to quickly create a new sample by copying information from the existing sample and making only the necessary modifications. To do so
- Go to Data Browser → Samples
- Locate the sample you would like to clone
- Right-click on the sample and choose "Clone" option
- Enter a unique name for the new sample in the popup window
- The Edit Sample window will appear. Make any necessary changes to the new sample by editing information in any of the five tabs:
- Sample
- Container
- Composition
- Sample Properties
- Additional Info
- Click "Save and Close" button to save the sample. The new sample will appear in the Samples section of the Data Browser.
| Tab | Name | Description |
|---|---|---|
| Sample | PI (required) | User selected PI (consists of PI(s) linked to the user |
| Display Name (required) | User specified name | |
| Physical State (required) | Solid-state NMR or solution NMR | |
| Preparation Date | Date when sample was prepared | |
| Preparation URL | Link to paper/protocols.io, etc. | |
| Description | Detailed description of the sample | |
| Notes | Other relevant sample information that the user wishes to enter | |
| Tags | List of user-defined tags that can be used for search and filter | |
| Container | Container Type | Information about rotor, tube or. other container |
| Notes | Other relevant information related to the sample container that the user wishes to enter | |
| Composition | Type | Sample Components |
| Name | User defined name of the pocmponent | |
| Role | Role of the component. The following options are available:
| |
| Quantity | User defined quantity of the component | |
| Unit | Concentration
Mass
Volume
| |
| Isotopic Labeling | Following options are available:
Additional user defined isotopic labeling details can be added/modified in the Actions | |
| Actions | Following actions are available:
| |
| Sample properties | pH Directly Measured | Indicate if the pH was measured directly in the NMR sample |
| pH value | pH value | |
| Aligned | Specify if the sample is under anisotropic alignment conditions (used for RDC or CSA measurements). | |
| Paramagnetic | Indicate if the sample contains paramagnetic center(s) | |
| Conductivity measured | Indicate whether the electrical conductivity of the sample was experimentally measured. If so, the value can be provided in μS/cm | |
| Sample volume | Sample volume in μL | |
| Ionic strength | ionic strength of the sample in mM | |
| Notes | Other relevant sample information that the user wishes to enter | |
| Additional Info | Storage/Handling details | User provided storage and handling conditions for the sample |
| Notes | Other relevant sample information that the user wishes to enter |